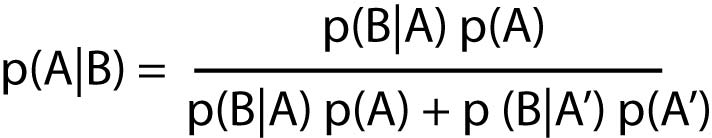Monte Carlo Methods
Named for the famous casino at Monte Carlo, these methods use randomization techniques to estimate a probability distribution in cases where it would be impractical to determine that distribution analytically.
Genetic Algorithms
Paul Lewis published a genetic algorithm for phylogenetic analysis using likelihood as the optimality criterion.
Genetic algorithms simulate evolutionary processes to solve analytical problems.
These algorithms create a population of possible solutions, and over many genertions (iterations) allow these solutions to "reproduce" and "mutate." The probability that any given solution will reproduce is a function of its optimality score, so in every generation the average score of the solutions in the population increases.
A genetic algorithm for phylogenetic analysis:
1) Create a matrix with a population of trees of different topologies. These could be random trees, or trees generated in some other way.
2) Calculate the likelihood of each tree (any other optimality criterion could also be used).
3) Populate a matrix with the next generation:
a) Copy the best tree to the next generation
b) Populate the remainder of the matrix with trees drawn from the previous generation by random sampling with replacement. The probability of drawing a given tree is a function of the likelihood of that tree (trees with higher likelihood are drawn more frequently). Each tree selected is changed randomly before being copied into the new matrix.
4) Return to (2)
This process is continued indefinitely. The longer the algorithm runs, the higher the probability of finding the globally optimal solution.
Bayesian Methods
Bayesian methods constitute a major branch of statistics, but are somewhat controversial.
Google's search engine is a Bayesian algorithm, so uppity Bayesians say that those who don't believe in Bayesian approaches shouldn't use Google.
Bayes' theorem on conditional probability
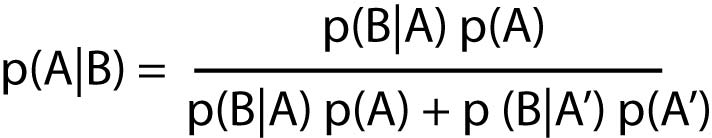
probability of event A, given event B
A' is the event "not A"
MrBayes - software implementing a Bayesian approach to phylogenetic analysis
Uses a MCMCMC algorithm: Metropolis-Hastings Coupled Markov Chain Monte Carlo
Markov Chain
Markov Process
A Markov process is a process in which the system can hold two or more states, there are probabilities of transition among those states, and there is no memory of what prior states have been held.
What future states can be held is a function only of what state the system is currently in.
A hidden Markov model models a Markov process, but the Markov states themselves are hidden, and one can only directly observe "emissions" from these states. The emission probabilities separate from the state-transition probabilities.
Metropolis-Hastings algorithm applied to phylogenetics
Represent a phylogenetic tree as a set of parameters for tree topology, branch length, model of sequence evolution, etc.
1) Start with test tree
2) Select a neighbor
3) Calculate ratio of probabilities of test tree and new tree
4) If ratio greater than or equal to 1, accept new tree
5) Else, draw a random number (between 0 and 1). If it is less than ratio, accept new tree.
Bollback, J. P. 2002. Bayesian model adequacy and choice in phylogenetics. Molecular Biology and Evolution 19: 1171-1180.
Huelsenbeck, J. P., F. Ronquist, R. Nielsen, and J. P. Bolback. 2001. Bayesian Inference of Phylogeny and Its Impact on Evolutionary Biology. Science 294: 2310-2314.
Larget, B., and D. L. Simon. 1999. Markov chain Monte Carlo algorithms for the Bayesian analysis of phylogenetic trees. Molecular Biology and Evolution 16: 750-759.
Lewis, P. O. 1998. A genetic algorithm for maximum-likelihood phylogeny inference using nucleotide sequence data. Molecular Biology and Evolution 15: 277-283.