 Charles B.
Fenster, Professor
Charles B.
Fenster, Professor
AFTER MANY PRODUCTIVE YEARS AT THE UNIVERSITY OF MARYLAND, CHARLIE FENSTER'S LAB HAS MOVED TO SOUTH DAKOTA STATE UNIVERSITY, BROOKINGS, SOUTH DAKOTA
WE WILL CONTINUE OUR RESEARCH AIMED AT ELUCIDATING EVOLUTIONARY PROCESS AND WILL NOW ALSO ADDRESS QUESTIONS IN CONSERVATION GENETICS AND SUSTAINABLE AGRICULTURE.
THANK YOU FOR YOUR INTEREST!
FOR MORE INFORMATION PLEASE CONNECT TO THE FOLLOWING WEBSITES
https://charlesbfenster.wordpress.com/
https://www.sdstate.edu/directory/charles-b-fenster
Welcome to the lab web page of Charles B. Fenster. Here you will find information on teaching and research with links to syllabi, publications, former students, postdoctoral fellows and collaborators. Please feel free to contact me via email if you have any questions regarding my teaching or research, or if you would like to work in the lab. High school students, undergraduates, graduate students and post Ph. D. postdocs and visitors are welcome.
Notable service:
AIBS BOARD (2011-present), AE Ecology and Evolution (2012- present), Chair Evolution 2013 Organizing Commitee, Chair and member UMD Senate Faculty Advisory Committee (2011 - 2013), Formerly Executive Vice President Society for the Study of Evolution, Formerly AE for International Journal of Plant Sciences, American Journal of Botany, Evolution
Note to Prospective Students and Post-Doctoral Fellows, Current Research Interests;
Expectations for Graduate Students
How to Write a Peer Review for a Journal Submission
Publications and PDF's (contact me if pdf is not present),
Link to Google Scholar Profile
Course Syllabi: Evolutionary Genetics
Current and Former Students and Post-doctoral Fellows
Non Technical Description of Research and Teaching
Academic Background:
- Stuyvesant High School
- Amherst College, BA
- University of Chicago, Ph. D. (Douglas W. Schemske, Ph.D. advisor)
- University of Toronto, Post-Doctoral Fellow (Kermit Ritland and Spencer C.H. Barrett, Postdoctoral Supervisors)
Academic Appointments:
- Full Professor, Department of Biology, The University of Maryland.
- Associate Professor, Department of Botany/Biology, The University of Maryland.
- Adjunct Professor, Xishuangbanna Tropical Botanical Garden, Chinese Academy of Sciences, Yunnan, China.
- Adjunct Professor, College of Life Sciences, Wuhan University, Wuhan, China.
- Professor, Botanisk Institutt, NTNU, 8/99- 8/2001.
- Assistant Professor, Depart of Botany, The University of Maryland.
Honors:
Executive Vice President Society for the Study of Evolution (2008-2010)
Board Member American Institute of Biological Sciences(2011-present)
Teaching: Teaching is as an important contribution to the maintenance of our democratic society. An informed public is critical to government that depends on participation. From a practical viewpoint, my teaching philosophy is to treat students as colleagues, where we endeavor to learn about the topic together. I use a Socratic approach as much as time will allow, and try to make the material relevant to the students by using appropriate examples. Throughout, I encourage a questioning attitude and I emphasize to students that scientists ask questions so that we might as well as get into the habit of doing so. In brief, my view of teaching is that it should accomplish three goals: 1) expose students to ideas, 2) encourage the growth of critical thinking, and 3) train students to express themselves cogently and accurately.
Recently taught courses:
Introductory
Genetics
Research: My work is broadly conceptually driven, focused on the elucidation of evolutionary process, using plants as model organisms. Because of the broad perspective of my research, I use a combination of approaches incorporating statistical, ecological, quantitative genetic and molecular marker based techniques to address the questions of my research program. While some of my work reflects greenhouse or laboratory based studies, for the most part my studies are conducted in the wild, where the connection between genotype and fitness, as mediated by natural selection can be quantified. Although much of my research focus is quantifying the input of genetic variation into the evolutionary process, albeit studied in the context of natural selection, there are many interesting details of natural selection that are relatively unknown, that if understood would provide much greater insight into the underpinnings of the evolutionary process. To examine the details of natural selection, I use flowers as models of adaptation. While the intellectual focus of the lab is on understanding fundamental issues of evolutionary biology, our work also provides insight for important concerns in conservation genetics and invasive species biology. I begin by summarizing ongoing projects and finish with a description of past projects, and questions that remain from those completed projects.
CURRENT PROJECTS
The Natural History of Mutations (with Matt Rutter, Associate Professor, College of Charleston, Stephen Wright, Associate Professor University of Toronto and Detlef Weigel, Director Max Planck Tübingen): Here we continue previous work on mutations (see below) by fully sequencing a number of Mutation Accumulation Lines in order to have a robust estimate of sequence level mutation rates and a comprehensive understanding of the relationship between types of mutations and fitness in the field.
Quantifying Mutation Parameters in the Wild (with Matt Rutter, Associate Professor, College of Charleston): The interplay between disorder and the generation of new order and complexity through mutation and selection acting on mutational variance is a fundamental feature of evolution. Yet how the effects of naturally occurring mutations both enable and constrain the evolution of diversity remains largely unexplored. The most fundamental aspects of the contribution of mutation to evolutionary processes involve the rate of generation of variation and the expression of those new mutations. Thus we are quantifying both the rate of spontaneous mutations affecting fitness and the distribution of mutation effects. This NSF funded research (to C. Fenster and M. Rutter) takes advantage of the macroscopic and sessile nature of the model organism, Arabidopsis thaliana, to quantify fitness variation of Mutation Accumulation Lines (developed by R. Shaw) in the wild and is being conducted with a postdoc, Matt Rutter, in collaboration with both Ruth and Frank Shaw.
To date we have planted the 100 Mutation Accumulation lines into an early successional field in Spring 2004 and 2005 and Fall 2004 and 2005 (total n = 40,000 plants at this point), representing the spring and winter-annual life histories found within the range of A. thaliana. We have also conducted a greenhouse study where we have measured mutation rates and effects for specific traits, floral, vegetative and physiological (delta C-13, with John McKay). To date, we have found that mutation effects appear to be symmetrically distributed about the original founder genotype, corresponding to Ruth Shaw’s earlier results, and contrary to the notion that mutations are uniformly deleterious. We will be able to examine the GxE effects corresponding to the performance of the Mutation Accumulation lines across temporal environments, providing not only an estimate of mutation effects on fitness sensitivity to the environment, but also how mutation inputs into the evolution of these GxE effects. We also have preliminary estimates of the whole genomic mutation rate for fitness using Peter Keightley’s maximum likelihood approach and Frank Shaw's MCMCML approach. We are currently collaborating with Jeff Conner’s lab (Angela Roles) to quantify mutation effects at the gene expression level, using microarray approaches.


Spring planting, A. thaliana, UVA Blandy Exptl. farm. A. thaliana several weeks before harvest.
Natural Selection- We have focused on those most perfect of adaptations, flowers, to quantify the role of natural selection in origin of floral diversity (see our 2004 article in Annual Review of Ecology, Evolution and Systematics). While my Mimulus genetic architecture papers (see below) demonstrate a signature of selection in mating system evolution, we are seeking to uncover the mechanisms of natural selection in much greater detail through work to quantify the role of pollinators as selective agents. Currently, with Michele Dudash, and Richard Reynolds, we, with funding from NSF, are quantifying the selective forces responsible for the origin and maintenance of pollination syndrome traits in three closely related species of Silene (Caryophyllaceae) (Dudash and Fenster 1997; Fenster and Dudash 2001). Because our work is conducted within a phylogenetic context, we can take advantage of the inherent power of comparative approaches. Thus some of our major goals include quantifying the important selective agents (pollinators) for the origin and maintenance of floral diversity among these three species and specifically what traits they select upon. We are using a combination of approaches to quantify selective pressures in two of the species, hummingbird pollinated S. virginica and nocturnal moth pollinated S. stellata, including cohort phenotypic selection analyses and experimental manipulation. In the former approach we are studying selection on about 1300 same-aged individuals over a five year period. Thus we hope to document phenotypic trade-offs between floral traits and vigor, a possible mechanism for the manifestation of stabilizing selection pressures. Particularly relevant to the above description of my research is our attempt to document the role of trait interaction in the evolution of pollination syndromes, exploring the utility of viewing the flower as a harmonious and coadaptively interacting organ, and the role of natural selection in generating this coadaptation from a phenotypic perspective. Recent observations with the moth pollinated species, S. stellata, indicate that a major pollinator, the noctuid moth Hadena ectypa, is also a major seed predator, since it oviposits one egg at the base of the petal with great frequency following taking nectar. Given that Silene is one of the largest genera of flower plants (over 700 species), with a range of breeding and pollination systems, and that Hadena is a seed parasite for many Silene species, as well as closely related outgroups, we believe that the Silene-Hadena interaction may be a model for studying the evolution of mutualisms, specifically in terms of the costs and benefits that dictate the evolutionary outcome of the interaction as either a mutualism or as parasitism (Kephart et al. 2006 New Phytologist).
Follow this link to see Hadena ectypa nectar and then lay an egg in a flower of Silene stellata
 |
 |
 |
| Hadena ectypa pollinating S. stellata | Ruby throated hummingbird pollinating S. virginica | Clear-wing hawk moth pollinating S. caroliniana (photo by M. Hood) |



Silene study sites: from left to right, first two near Mountain Lake Biological Station, SW Virginia, last, Great Falls, Potomac River near Washington DC.
Additional ongoing projects focused on pollination and mating-system evolution: With Silvana Marten-Rodriguez and Liz Zimmer, we are using observational, experimental manipulations as well as comparative approaches to understandi the selective forces that underlay the adaptive radiation of Gesnerieae in the Caribbean (see link to Silvana's web site). I also have overseas collaborations with Johanna Maad (Uppsala University) and Scott Armbruster (NTNU and Portsmouth University) to study the adaptive significance, if any, of the floral size polymorphism exhibited by Campanula rotundifolia in contrasting low and high elevational sites in near-arctic environments found in the valleys and mountain tops of Norway ( Norwegian Research Council to J. Maad). I have an additional collaboration with Shuang-Quan Huang (Wuhan University) to understand the adaptive significance of the great diversity of floral form exhibited by Pedicularis in the Himalayas (Chinese National Science Foundation to S-Q Huang).
 |
 |
| Campanula rotundifolia at approximately 1000 m, near Oppdahl, Norway | Hummingbird visit to Rhytidophyllum vernicosum, Dominican Republic(photo by S. Martén-Rodriguez) |
The Evolutionary Significance of Epistasis: Much of my research focus has been to explore the relevance of limited gene flow to the evolutionary process by focusing on estimating the degree to which populations of C. fasciculata are inbred and the extent to which epistasis contributes to population genetic differentiation. Inbreeding and epistasis are of major interest because they distinguish two of the major models of evolutionary process produced early on in the neo-darwinian synthesis (see Fenster et al. 1997 for discussion) (NSF to C. Fenster). If populations are indeed small, then inbreeding effects should be apparent and populations will evolve with the genetic variation present, with different populations reaching similar degrees of adaptation to the environment but through different genetic mechanisms. These indeed were the major conclusions of the epistasis and inbreeding work, that is, populations were so inbred that F1 hybrids between ecotypically differentiated populations outperformed the parents (Conservation Biology 2000), and epistasis featured prominently in the genetic differentiation of populations (Evolution 1999, 2000ab, 2001). However, in a recently published paper (Evolution 2006), we observed that much later F6 generation hybrids recovered fitness in a manner consistent with the contribution of epistasis to the de novo synthesis of beneficial coadaptive gene complexes. Thus, to date, our research has demonstrated that epistasis figures prominently in the evolution of populations, both in terms of population differentiation and potentially to the introduction of novel gene combinations important to selection response through hybridization or long gene flow. Currently, my lab is quantifying in greater detail the role of epistasis in population differentiation of C. fasciculata by using QTL (=marker based approaches) methodologies to quantify the genetic basis of ecotypic differentiation using microsatellite and AFLP markers (see our paper, Erickson et al. 2004, in Molecular Ecology to view my thoughts regarding QTL approaches). The current QTL work will reveal greater details of the underlying genetic basis of fitness differentiation in natural populations, confirming and expanding interpretation of previous results. Since epistasis figures prominently in population differentiation (at least in C. fasciculata), one can ask how quickly these genetic background effects evolve? If evolution frequently represents fixation of alleles of relatively small effect at many loci, then how can small populations evolve? These questions and many others in evolutionary biology ultimately hinge on our understanding of mutation parameters (see above).
 |
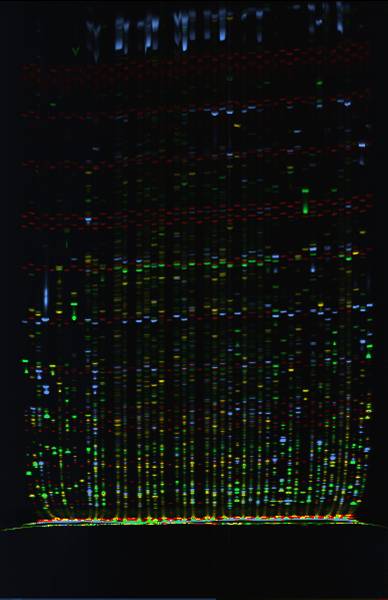 |
| David Erickson at our study site, Konza Prairie, near Manhattan Kansas | Gel of AFLP bands used in our QTL analyses to quantify
the genetic architecture of ecotypic differentiation |
SUMMARY OF MAJOR PAST PROJECTS:
Gene Flow- Because so much in evolution is context dependent, much of my research has explored the context dependency of the evolutionary process. I initially explored context dependency by quantifying the size of populations in a genetic sense because whether evolutionary process is embedded within small or large populations determines the relevancy of many evolutionary models. In studies initiated while a graduate student with Douglas Schemske (NSF to D. Schemske and C. Fenster), we demonstrated in three Evolution papers (1991ab, 2003) that gene flow is very limited in a native North American legume (Chamaecrista fasciculata) and the level of gene flow corresponded precisely with the pattern of genetic variation of neutral allozyme markers. Genetic divergence within this species occurs within a context of an isolation by distance model of gene flow. These results led to several further questions. If populations of C. fasciculata are relatively genetically isolated from one another, then how does this affect the spread of advantageous alleles? Does population genetic structuring merely retard adaptive evolution, or does it fundamentally change the way it occurs? For example, genetic background effects may be very important with selection acting not only on individual alleles but how those alleles genetically interact with genetic variation at other loci (epistasis). These types of questions can be addressed by close examination of the genetic architecture (number of loci, mode of expression, degree of pleiotropy etc.) underlying population and species differentiation. Other open questions would be to extend this work to a Meta-Population perspective using chloroplast and microsatellite markers, which have been developed in the lab.

Chamaecrista fasciculata
Mating-System Evolution- In my first forays into quantifying genetic architecture, as a postdoc in Kermit Ritland’s and Spencer Barrett’s labs at the University of Toronto, I explored the genetic architecture of mating system evolution (the evolution of small flowered selfing species from large flowered outcrossing species) in order to determine whether evolution is either mutation limited or selection limited (NSERC funding to K. Ritland and S. Barrett). This was accomplished by crossing programs to quantify whether the divergence of floral structure represented the fixation of alleles of small effect at many loci or the fixation of one or a few major effect alleles, conforming to the selection and mutation limited models of evolutionary process, respectively. This work with Mimulus (Fenster and Ritland 1994; Carr and Fenster 1994; Fenster et al. 1995; Fenster and Carr 1997) conformed to the selection limited model of evolutionary mechanisms and was later confirmed by QTL studies conducted by Willis and colleagues. However, our studies of Eichhornia paniculata (Fenster and Barrett 1994) demonstrated that the breakdown of tristyly and subsequent evolution of selfing in this outcrossing species likely reflected the initial action of a single locus with large effect, elevating one of the anthers to be proximate to the stigma, facilitating automatic or simultaneous selfing. It is likely that adaptive evolution reflects a range of genetic architectures, depending on the traits, organisms, and fitness topography generating selective pressures.

Top Row: Parental Generation (Mimmulus micranthus
x M. guttatus); Middle Row: F1; Bottom Row: F2
Charles Fenster, Matt Rutter and Leo (at
end of A. thaliana harvest)
===================================================================================================

Charlie Fenster, Matt Rutter and Leo after a successful Arabidopsis harvest
email: cfenster@umd.edu